3-Package Functions with Examples
Source:vignettes/3-package-functions-with-examples.Rmd
3-package-functions-with-examples.Rmddata
Demo Data
The demo data includes the list of indicators/criteria in different langues
readxl::read_excel( system.file("data-demo/indicator_criteria.xlsx", package = "VulnerabilityScoreCalibration"),
sheet = "indicator") |>
dplyr::filter( language == "English (en)") |>
dplyr::select( dimension, indicator, indicator_hint)
#> # A tibble: 22 × 3
#> dimension indicator indicator_hint
#> <chr> <chr> <chr>
#> 1 Demographic Profile Presence of specific (at-risk) profiles i… Includes: pre…
#> 2 Demographic Profile Language ability of the head of household Head of house…
#> 3 Demographic profile Presence of health-based risk profiles At least one …
#> 4 Demographic Profile Dependency ratio Number of mem…
#> 5 Basic Needs Access to health services when needed (in… Was or was no…
#> 6 Basic Needs Access to meals per day in the last week Evaluates num…
#> 7 Basic Needs Access to drinking water NA
#> 8 Basic Needs Access to basic hygiene items in the last… NA
#> 9 Living conditions Access to bathroom (private or shared) Considering w…
#> 10 Living conditions Type of housing/shelter Includes: own…
#> # ℹ 12 more rowsThe following dataset has been anonymized from one of the field implementation.
kobodata <- system.file("data-demo/quadra_data.xlsx", package = "VulnerabilityScoreCalibration")
koboform <- system.file("data-demo/quadra_form.xlsx", package = "VulnerabilityScoreCalibration")Quadratic Voting
quadratic_prepare
indicator <- system.file("data-demo/indicator_criteria.xlsx", package = "VulnerabilityScoreCalibration")
quadratic_prepare()
#> NULLquadratic_review
# kobodata <- here::here("", "quadra_data.xlsx")
# koboform <- here::here("", "survey_quadraticvoting_CBI_Indicators.xlsx")
kobodata <- system.file("data-demo/quadra_data.xlsx", package = "VulnerabilityScoreCalibration")
koboform <- system.file("data-demo/quadra_form.xlsx", package = "VulnerabilityScoreCalibration")
## Run the process
result <- quadratic_review(kobodata, koboform)
#> Joining with `by = join_by(groups.positions.pos_name)`
#> Joining with `by = join_by(groups.positions.pos_name)`
## Review output
result[["topic_prioritisation"]]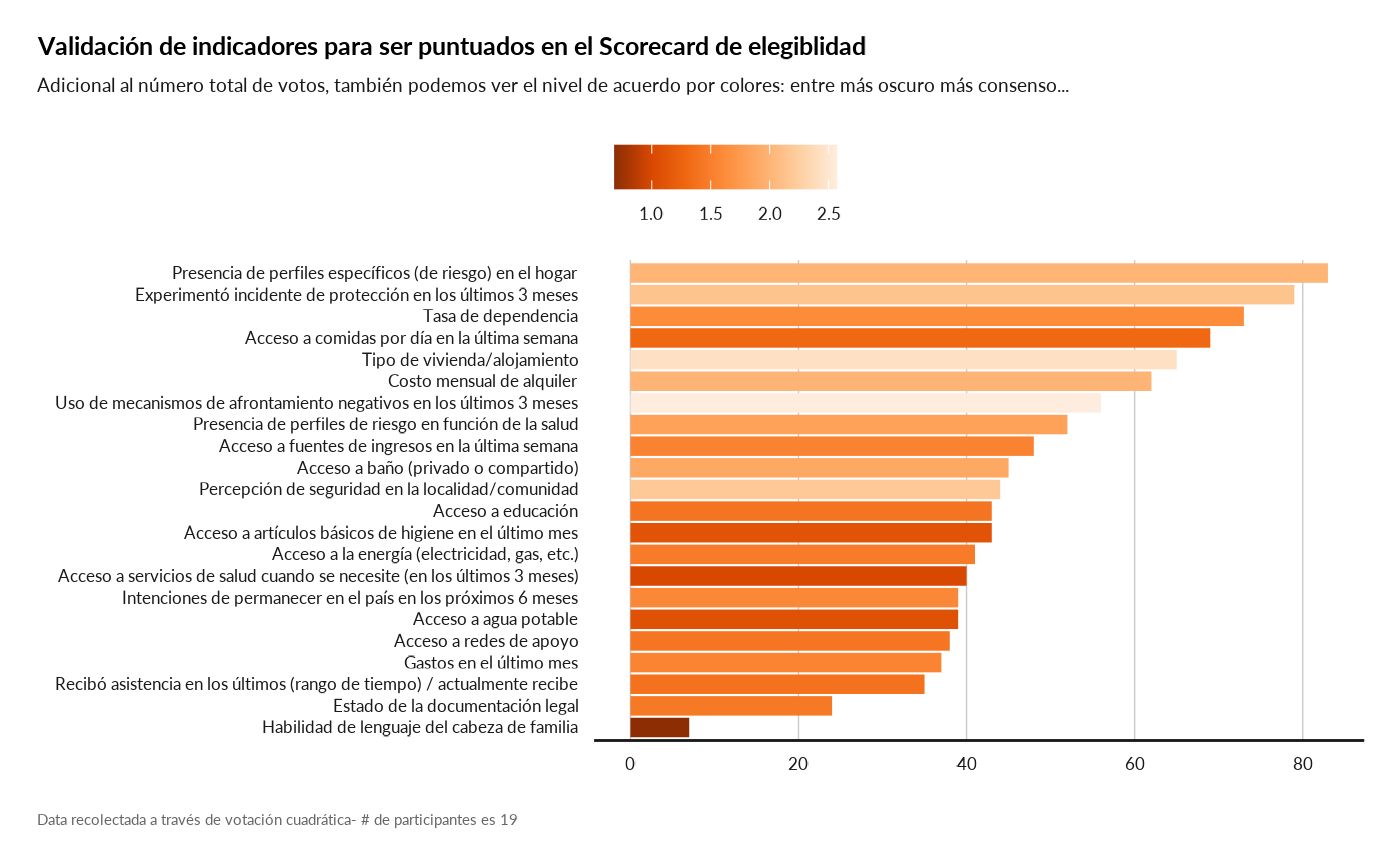
result[["vote_dispersion"]]
#> Picking joint bandwidth of 0.681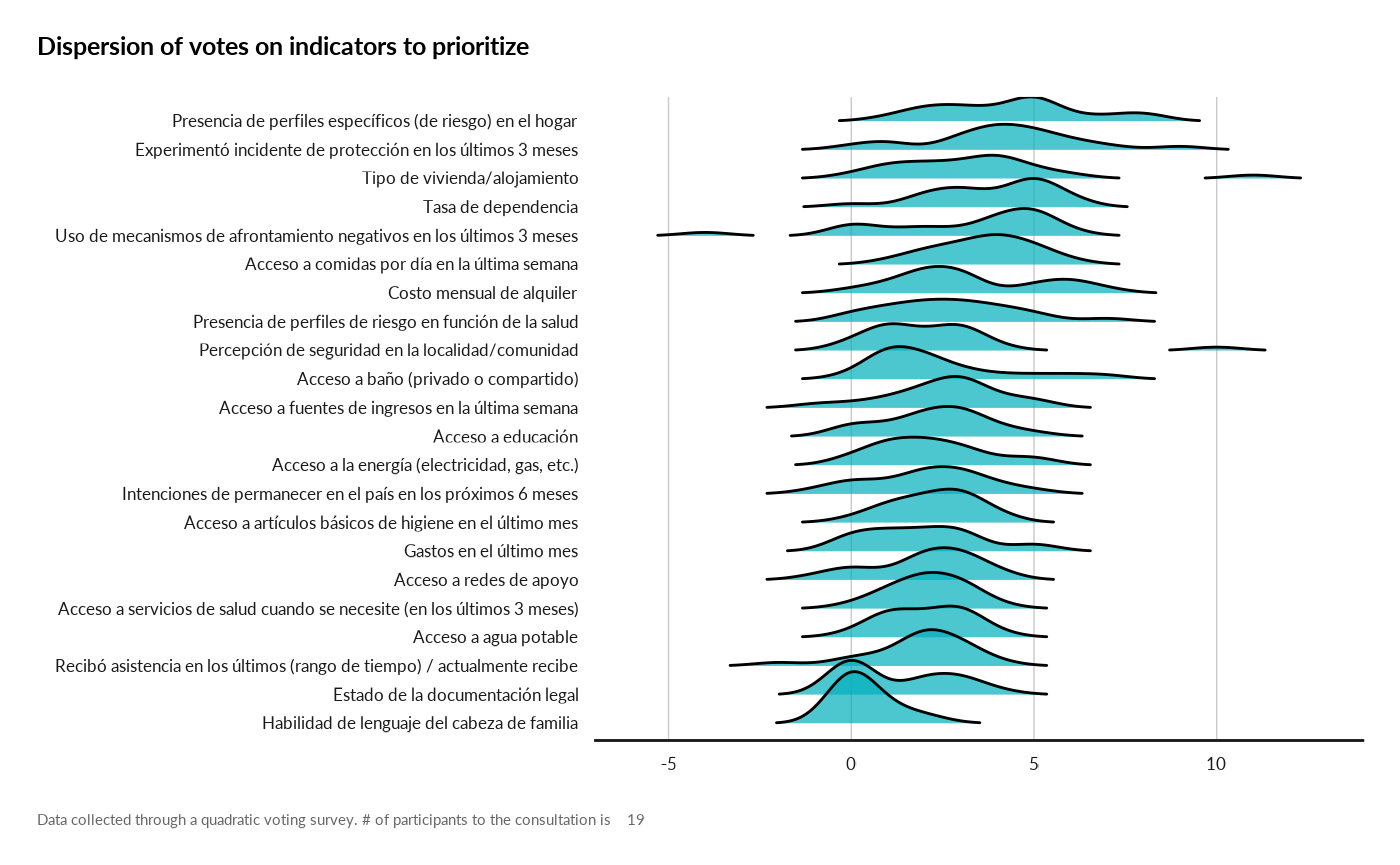
result[["individual_prioritisation"]]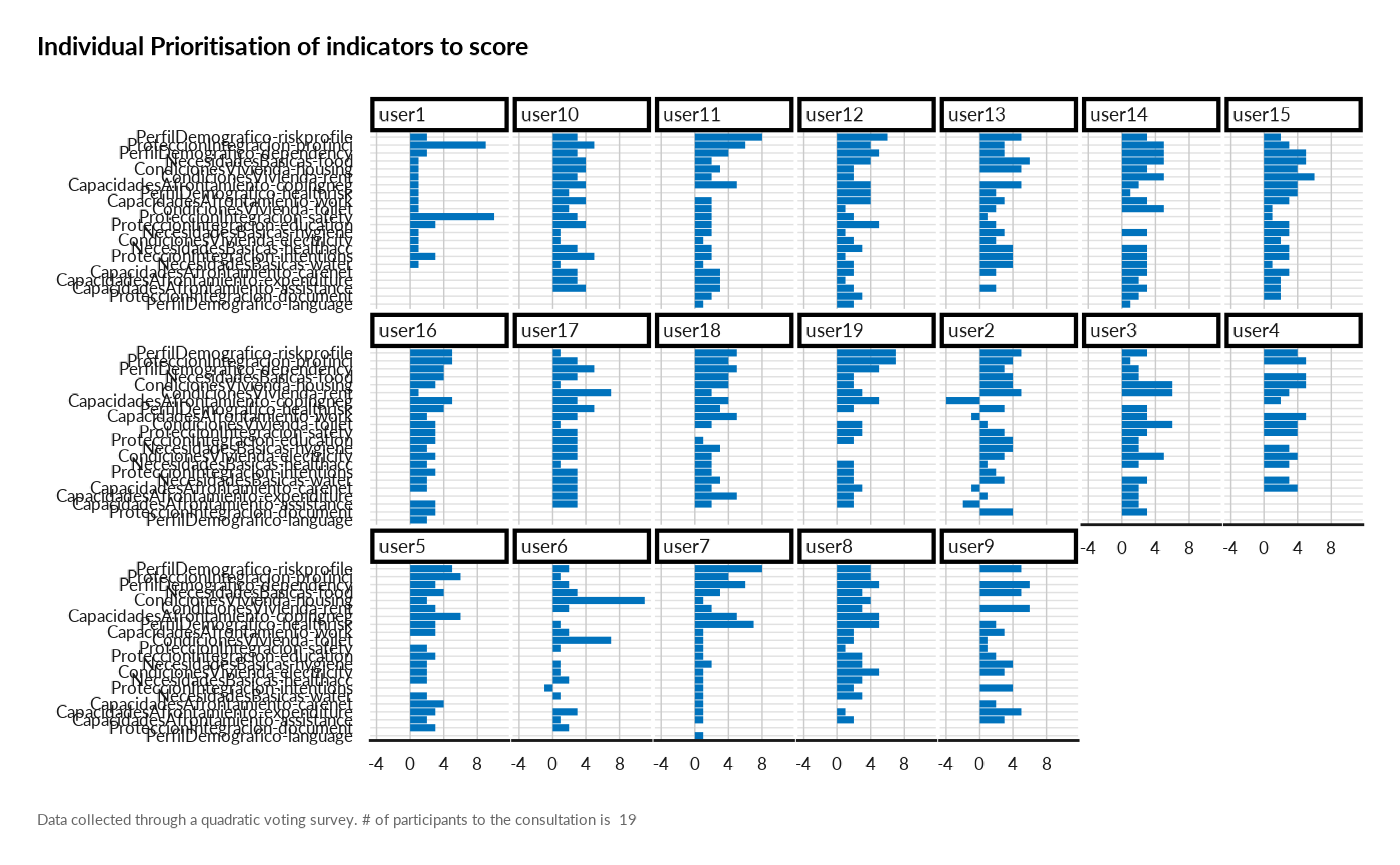
Conjoint Analysis
conjoint_prepare
# indicator <- system.file("data-demo/indicator_criteria.xlsx", package = "VulnerabilityScoreCalibration")
# opts <- read_excel("cja_opts_SAL.xlsx")
#
# conjoint_prepare( opts = opts,
# language = "Spanish (es)",
# form_title = "Actividad #2: Calificación de perfiles de vulnerabilidad - El Salvador, 23 de marzo, 2023",
# id_string = "vulnerability_rating",
# outdir = "",
# outfile = "form.xlsx" )conjoint_review
kobodata <- system.file("data-demo/conjoint_data.xlsx", package = "VulnerabilityScoreCalibration")
koboform <- system.file("data-demo/conjoint_form.xlsx", package = "VulnerabilityScoreCalibration")
cj <- conjoint_review(kobodata, koboform)
#> New names:
#> Joining with `by = join_by(dim, feature)`
#> • `` -> `...1`
#> • `` -> `...2`
#> • `` -> `...3`
#> Warning: There were 12 warnings in `dplyr::mutate()`.
#> The first warning was:
#> ℹ In argument: `margins = list(cregg::mm(data, as.formula(formula), id =
#> ~email))`.
#> ℹ In row 1.
#> Caused by warning in `logLik.svyglm()`:
#> ! svyglm not fitted by maximum likelihood.
#> ℹ Run `dplyr::last_dplyr_warnings()` to see the 11 remaining warnings.
#> Warning: Returning more (or less) than 1 row per `summarise()` group was deprecated in
#> dplyr 1.1.0.
#> ℹ Please use `reframe()` instead.
#> ℹ When switching from `summarise()` to `reframe()`, remember that `reframe()`
#> always returns an ungrouped data frame and adjust accordingly.
#> ℹ The deprecated feature was likely used in the VulnerabilityScoreCalibration
#> package.
#> Please report the issue at
#> <https://github.com/unhcr-americas/VulnerabilityScoreCalibration/issues>.
#> This warning is displayed once every 8 hours.
#> Call `lifecycle::last_lifecycle_warnings()` to see where this warning was
#> generated.
#> Loading required package: patchwork
cj[["data_quality"]]
conjoint_plot_point
kobodata <- system.file("data-demo/conjoint_data.xlsx", package = "VulnerabilityScoreCalibration")
koboform <- system.file("data-demo/conjoint_form.xlsx", package = "VulnerabilityScoreCalibration")
cj <- conjoint_review(kobodata, koboform)
#> New names:
#> Joining with `by = join_by(dim, feature)`
#> • `` -> `...1`
#> • `` -> `...2`
#> • `` -> `...3`
#> Warning: There were 12 warnings in `dplyr::mutate()`.
#> The first warning was:
#> ℹ In argument: `margins = list(cregg::mm(data, as.formula(formula), id =
#> ~email))`.
#> ℹ In row 1.
#> Caused by warning in `logLik.svyglm()`:
#> ! svyglm not fitted by maximum likelihood.
#> ℹ Run `dplyr::last_dplyr_warnings()` to see the 11 remaining warnings.
conjoint_plot_point( as.data.frame(cj[["cjdata"]][1,][["margins"]])) +
ggplot2::labs( subtitle = "Margins)")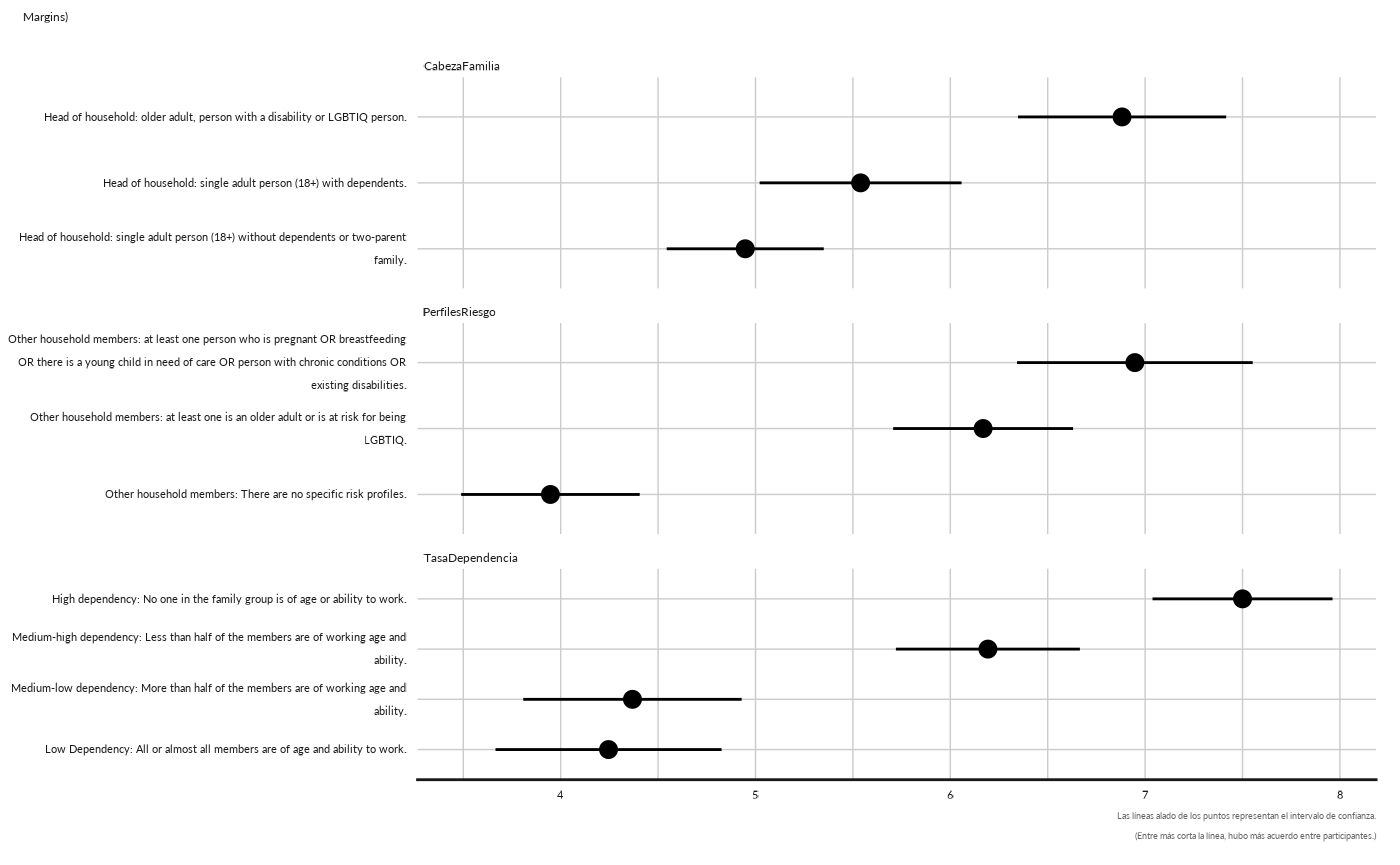
conjoint_plot_point( as.data.frame(cj[["cjdata"]][1,][["amces"]])) +
ggplot2::labs( subtitle = "Average Marginal Component Effects (AMCEs)")
#> Warning: Removed 1 rows containing missing values (`geom_segment()`).
#> Warning: Removed 1 rows containing missing values (`geom_segment()`).
#> Removed 1 rows containing missing values (`geom_segment()`).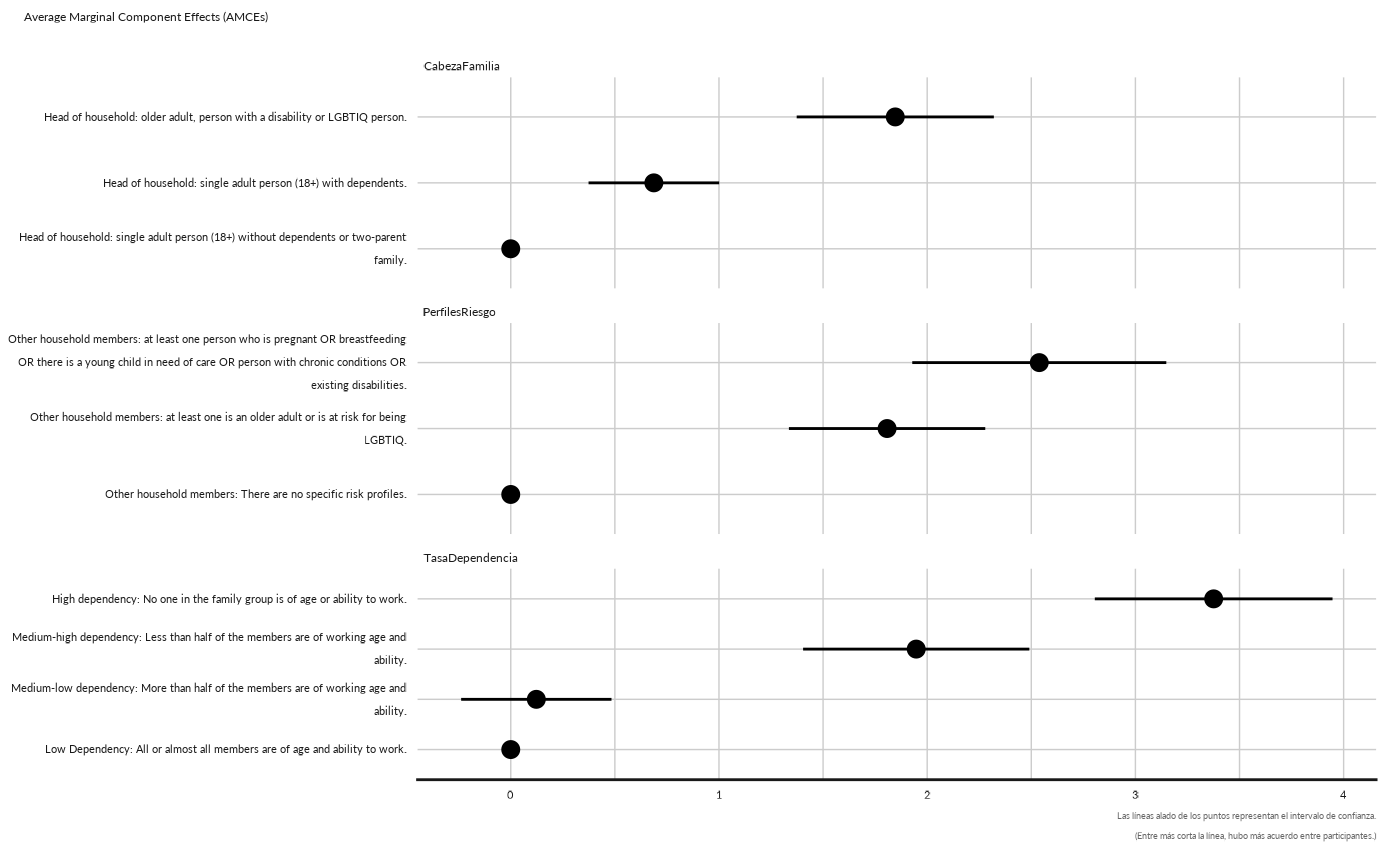
conjoint_plot_point( as.data.frame(cj[["cjdata"]][1,][["importance"]])) +
ggplot2::labs( subtitle = "Importance")
#> Warning: Removed 1 rows containing missing values (`geom_segment()`).
#> Removed 1 rows containing missing values (`geom_segment()`).
#> Removed 1 rows containing missing values (`geom_segment()`).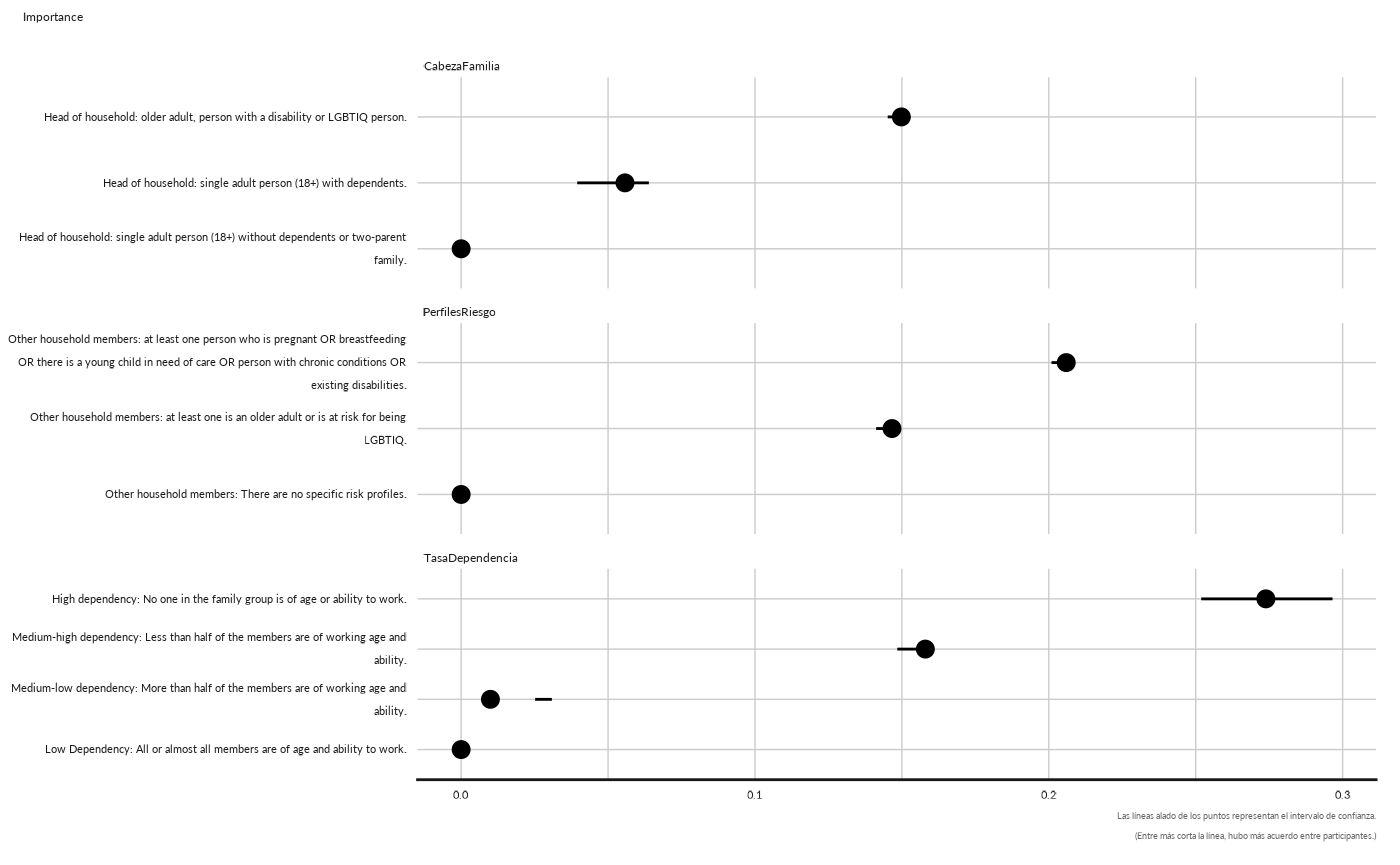
conjoint_plot_bar - Average Marginal Component Effects (AMCEs)
kobodata <- system.file("data-demo/conjoint_data.xlsx", package = "VulnerabilityScoreCalibration")
koboform <- system.file("data-demo/conjoint_form.xlsx", package = "VulnerabilityScoreCalibration")
cj <- conjoint_review(kobodata, koboform)
#> New names:
#> Joining with `by = join_by(dim, feature)`
#> • `` -> `...1`
#> • `` -> `...2`
#> • `` -> `...3`
#> Warning: There were 12 warnings in `dplyr::mutate()`.
#> The first warning was:
#> ℹ In argument: `margins = list(cregg::mm(data, as.formula(formula), id =
#> ~email))`.
#> ℹ In row 1.
#> Caused by warning in `logLik.svyglm()`:
#> ! svyglm not fitted by maximum likelihood.
#> ℹ Run `dplyr::last_dplyr_warnings()` to see the 11 remaining warnings.
## Plot AMCES as bar for dimension 2
conjoint_plot_bar( as.data.frame(cj[["cjdata"]][2,][["amces"]])) +
ggplot2::labs( subtitle = "Average Marginal Component Effects (AMCEs)")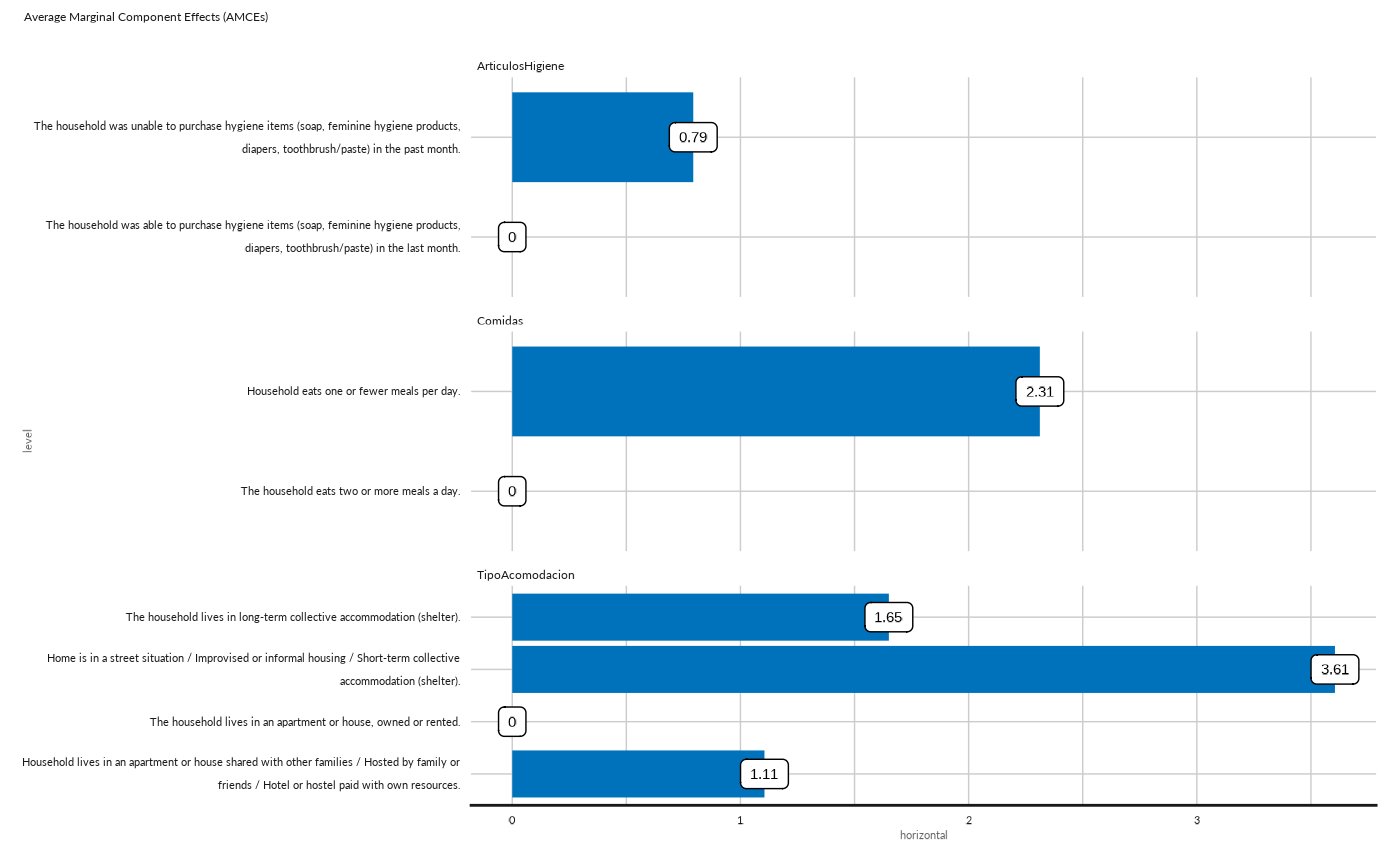
## Plot importance as bar for dimension 2
conjoint_plot_bar( as.data.frame(cj[["cjdata"]][2,][["importance"]])) +
ggplot2::labs( subtitle = "Importance")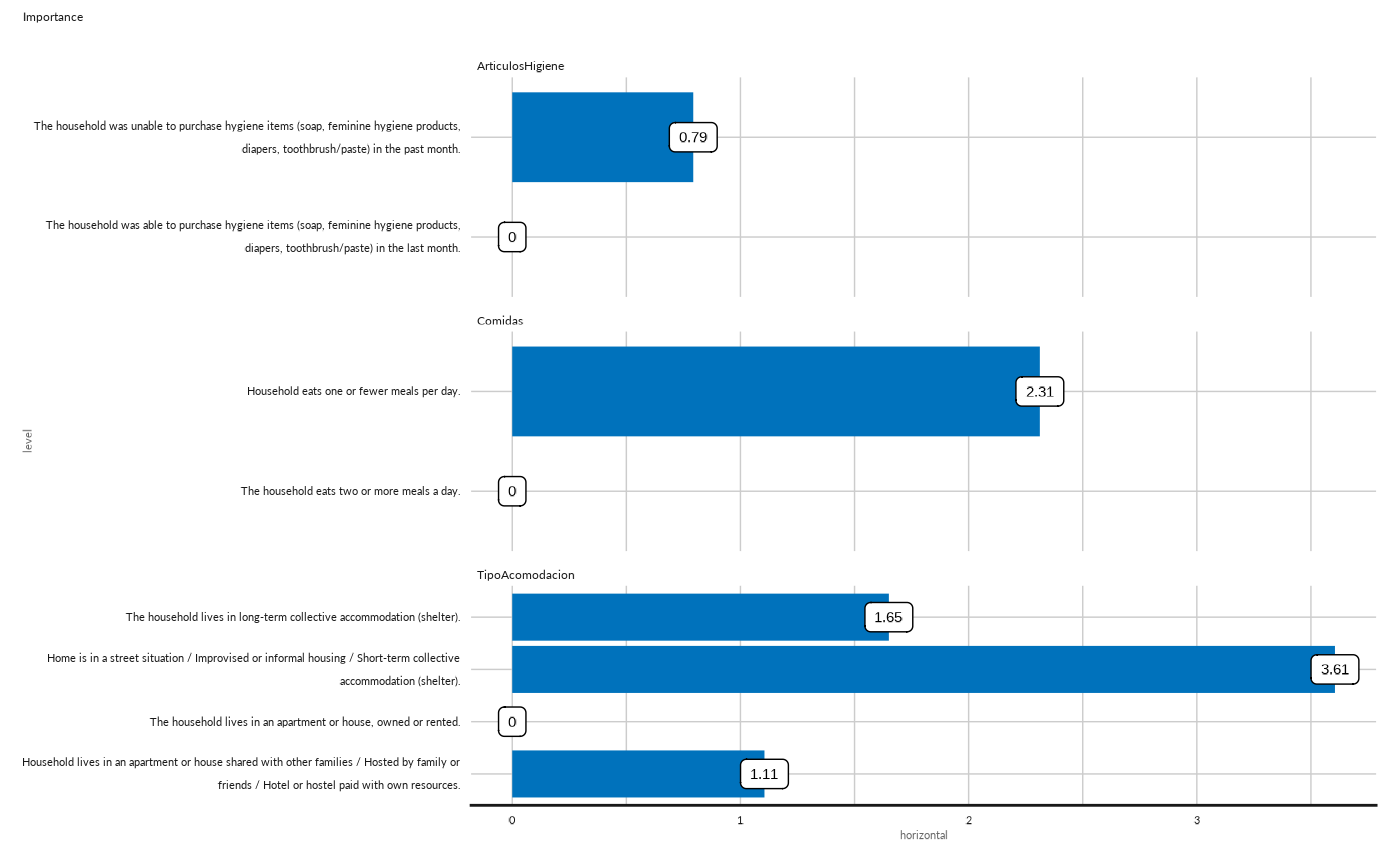
conjoint_walk - Summary by dimension
kobodata <- system.file("data-demo/conjoint_data.xlsx", package = "VulnerabilityScoreCalibration")
koboform <- system.file("data-demo/conjoint_form.xlsx", package = "VulnerabilityScoreCalibration")
cj <- conjoint_review(kobodata, koboform)
#> New names:
#> Joining with `by = join_by(dim, feature)`
#> • `` -> `...1`
#> • `` -> `...2`
#> • `` -> `...3`
#> Warning: There were 12 warnings in `dplyr::mutate()`.
#> The first warning was:
#> ℹ In argument: `margins = list(cregg::mm(data, as.formula(formula), id =
#> ~email))`.
#> ℹ In row 1.
#> Caused by warning in `logLik.svyglm()`:
#> ! svyglm not fitted by maximum likelihood.
#> ℹ Run `dplyr::last_dplyr_warnings()` to see the 11 remaining warnings.
cjdata <- cj[["cjdata"]]
## Get a summary of all dimensions
purrr::pwalk(cjdata, conjoint_walk)
#> ---
#>
#> PerfilDemografico
#> ---
#>
#> ## Average Marginal Component Effects (AMCEs) - Bar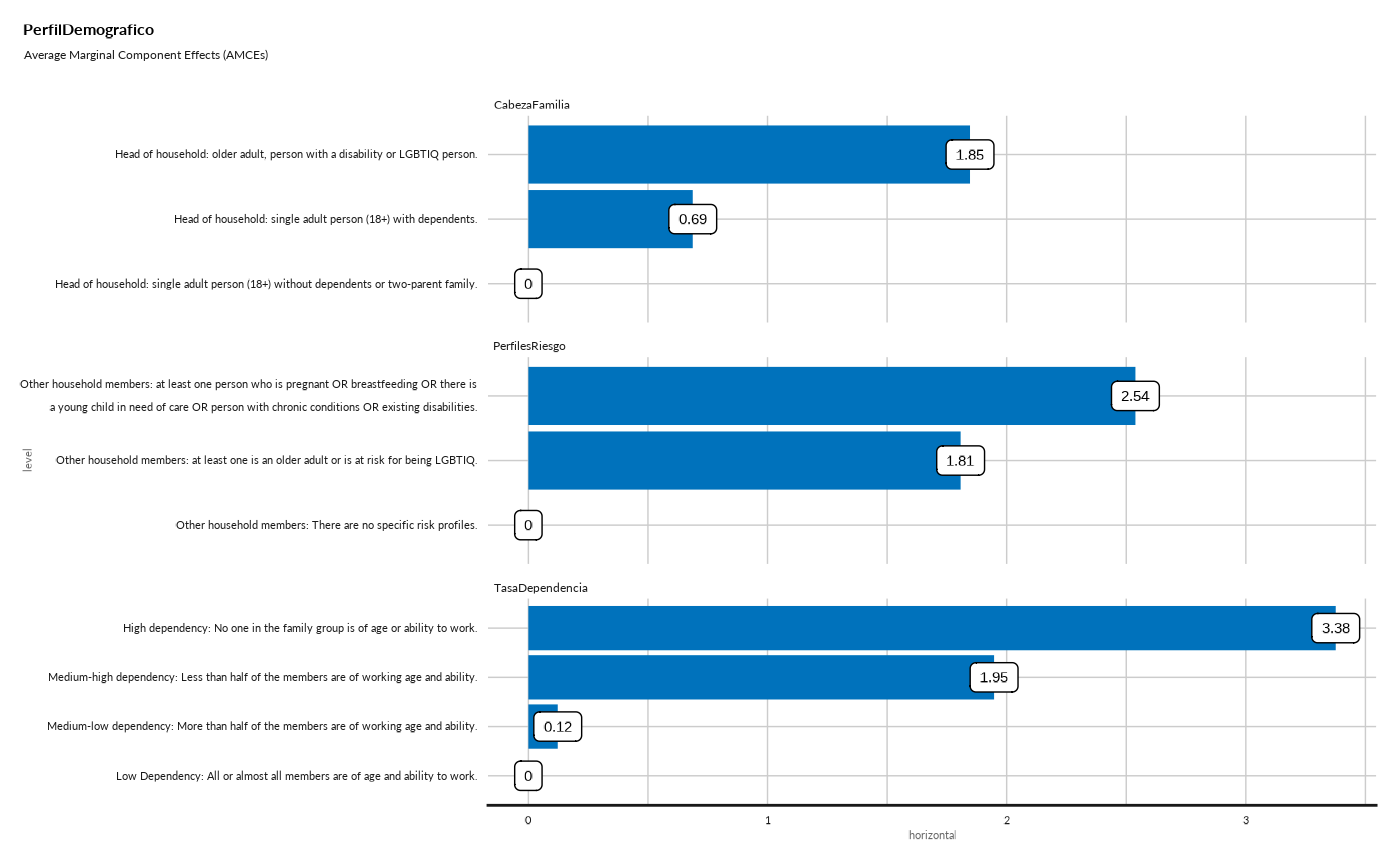
#>
#>
#> ## Average Marginal Component Effects (AMCEs) - Point
#> Warning: Removed 1 rows containing missing values (`geom_segment()`).
#> Warning: Removed 1 rows containing missing values (`geom_segment()`).
#> Removed 1 rows containing missing values (`geom_segment()`).
#>
#>
#> ## Marginal Means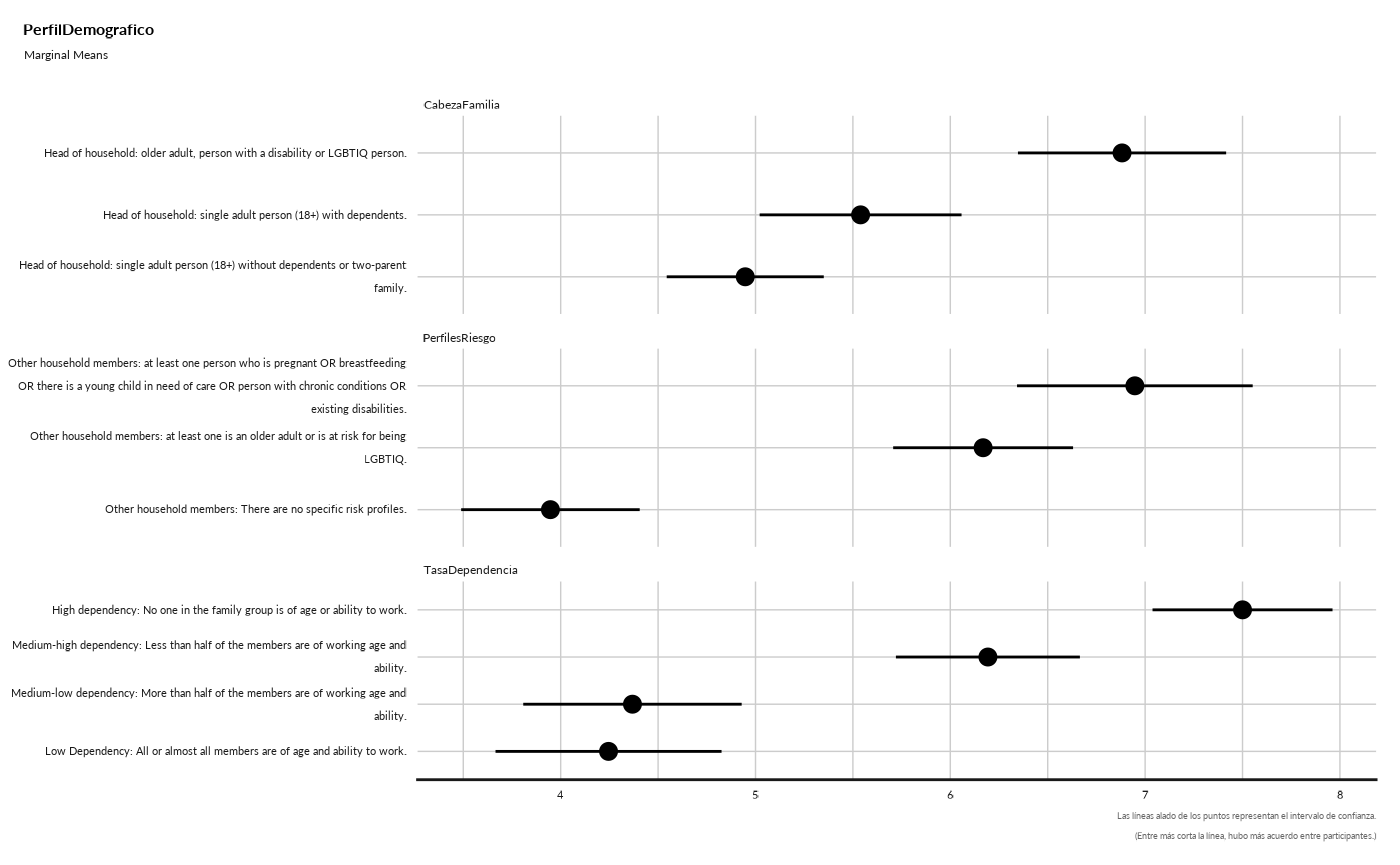
#>
#>
#> ## Importance Weights
#> Warning: Removed 1 rows containing missing values (`geom_segment()`).
#> Removed 1 rows containing missing values (`geom_segment()`).
#> Removed 1 rows containing missing values (`geom_segment()`).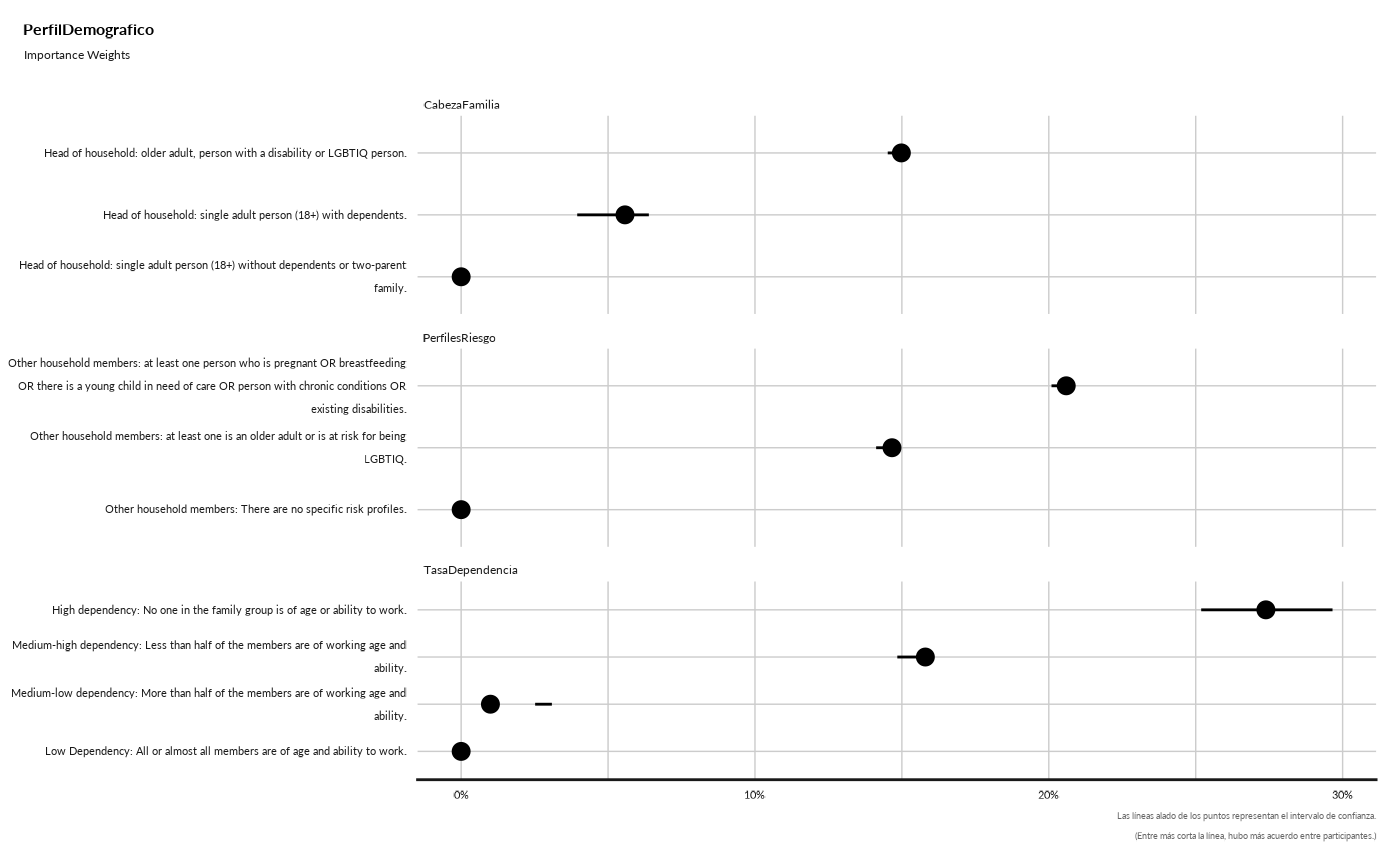
#>
#>
#> ---
#>
#> NecesidadesBasicas
#> ---
#>
#> ## Average Marginal Component Effects (AMCEs) - Bar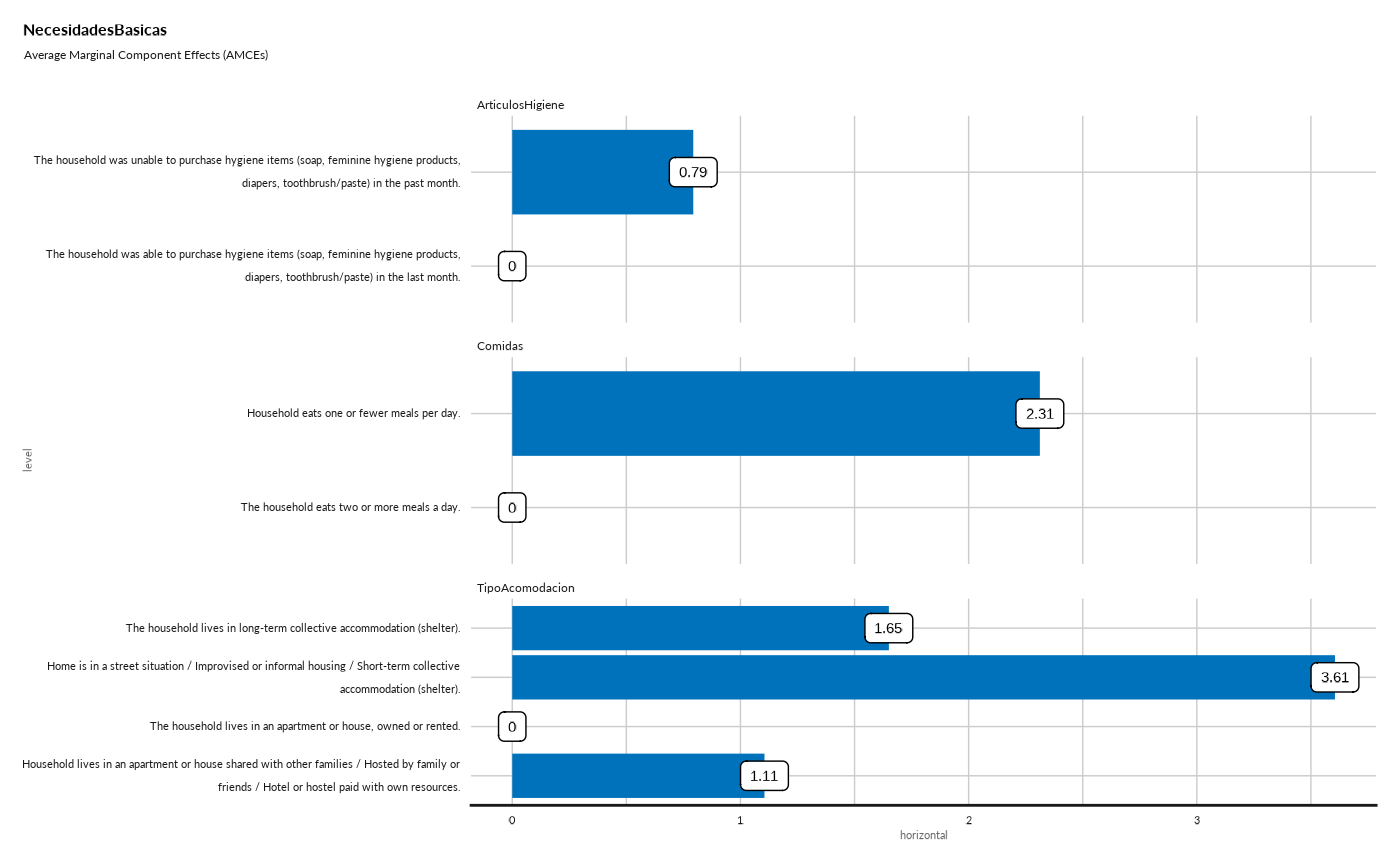
#>
#>
#> ## Average Marginal Component Effects (AMCEs) - Point
#> Warning: Removed 1 rows containing missing values (`geom_segment()`).
#> Removed 1 rows containing missing values (`geom_segment()`).
#> Removed 1 rows containing missing values (`geom_segment()`).
#>
#>
#> ## Marginal Means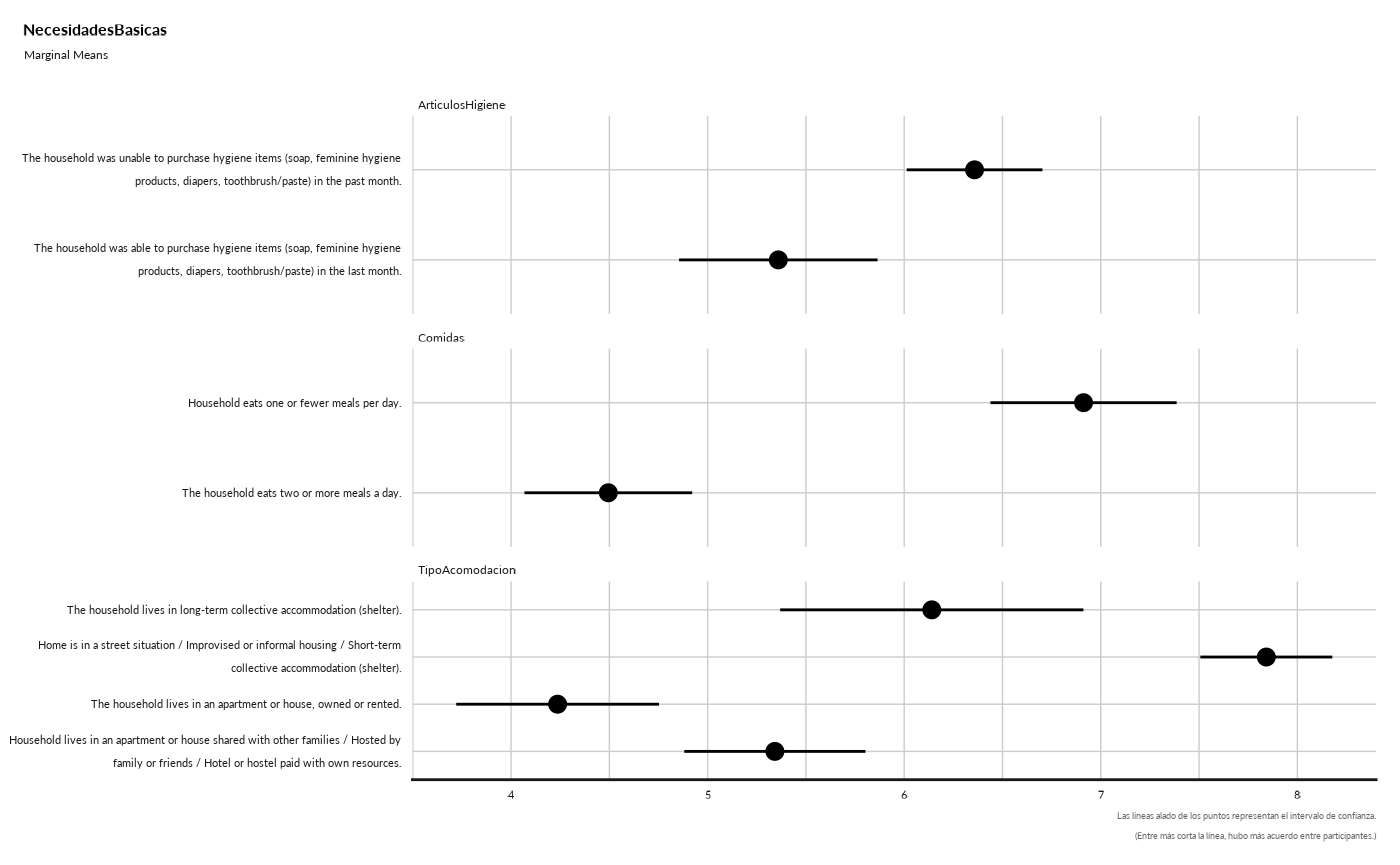
#>
#>
#> ## Importance Weights
#> Warning: Removed 1 rows containing missing values (`geom_segment()`).
#> Removed 1 rows containing missing values (`geom_segment()`).
#> Removed 1 rows containing missing values (`geom_segment()`).
#>
#>
#> ---
#>
#> AfrontamientoProteccion
#> ---
#>
#> ## Average Marginal Component Effects (AMCEs) - Bar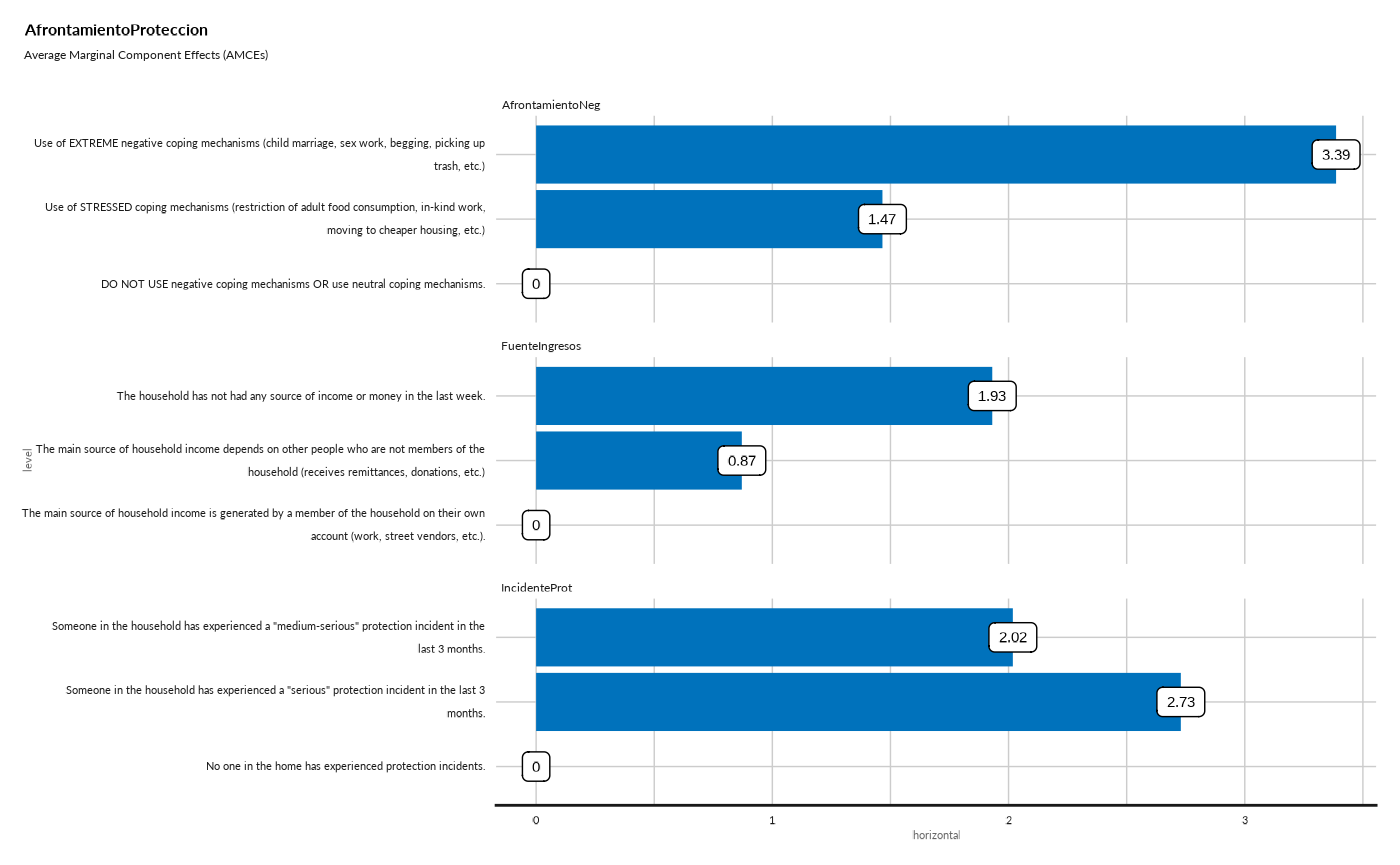
#>
#>
#> ## Average Marginal Component Effects (AMCEs) - Point
#> Warning: Removed 1 rows containing missing values (`geom_segment()`).
#> Removed 1 rows containing missing values (`geom_segment()`).
#> Removed 1 rows containing missing values (`geom_segment()`).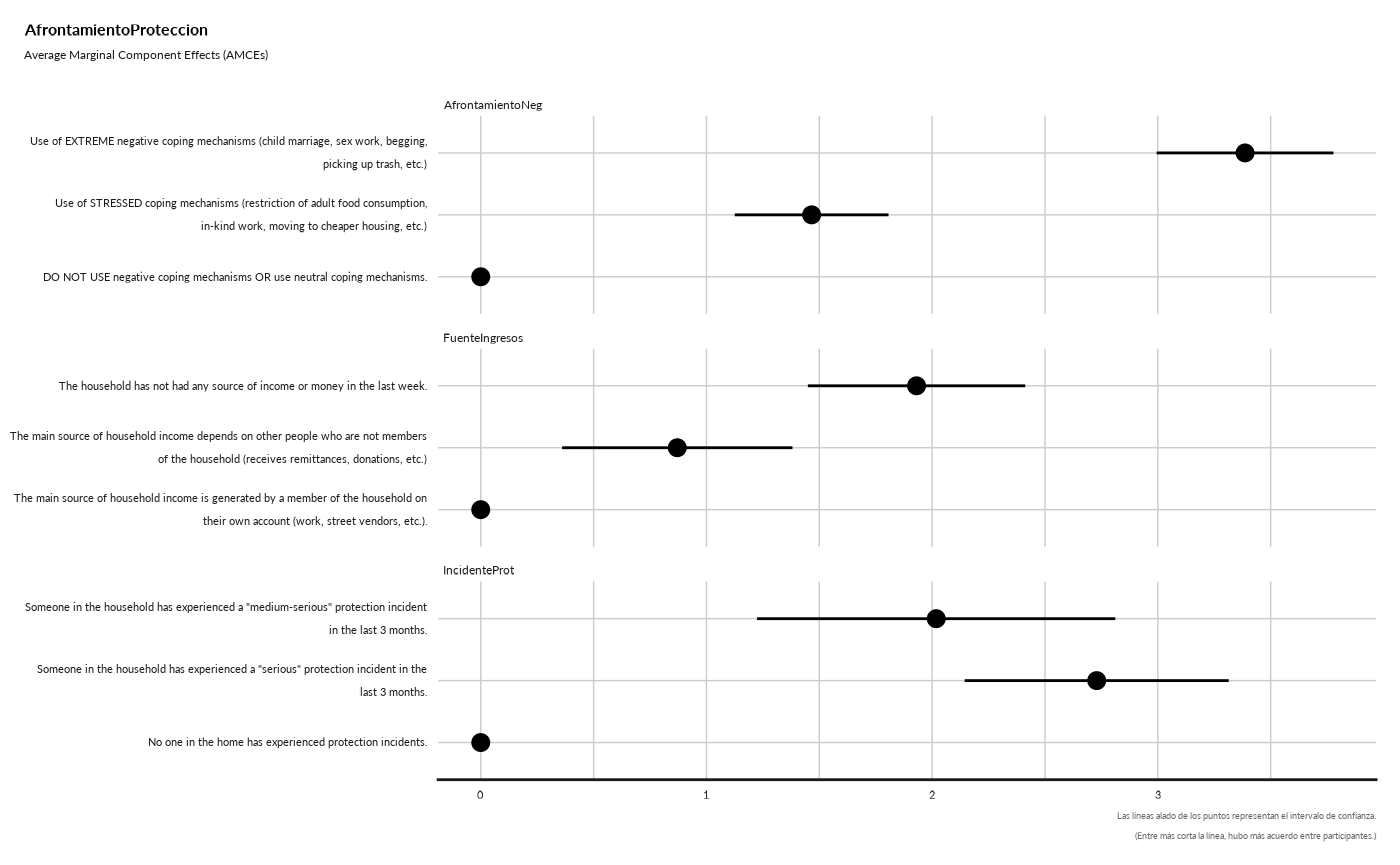
#>
#>
#> ## Marginal Means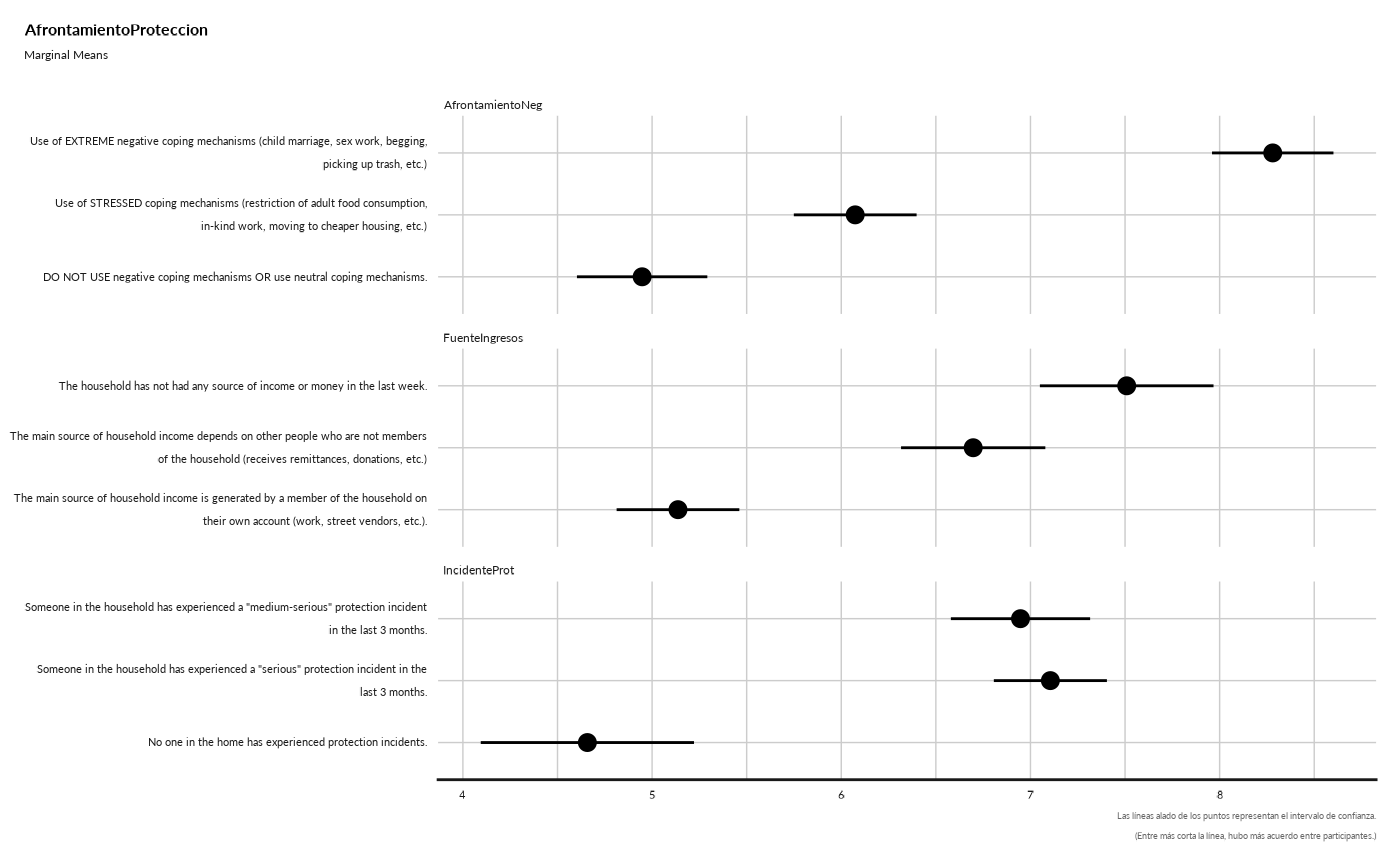
#>
#>
#> ## Importance Weights
#> Warning: Removed 1 rows containing missing values (`geom_segment()`).
#> Removed 1 rows containing missing values (`geom_segment()`).
#> Removed 1 rows containing missing values (`geom_segment()`).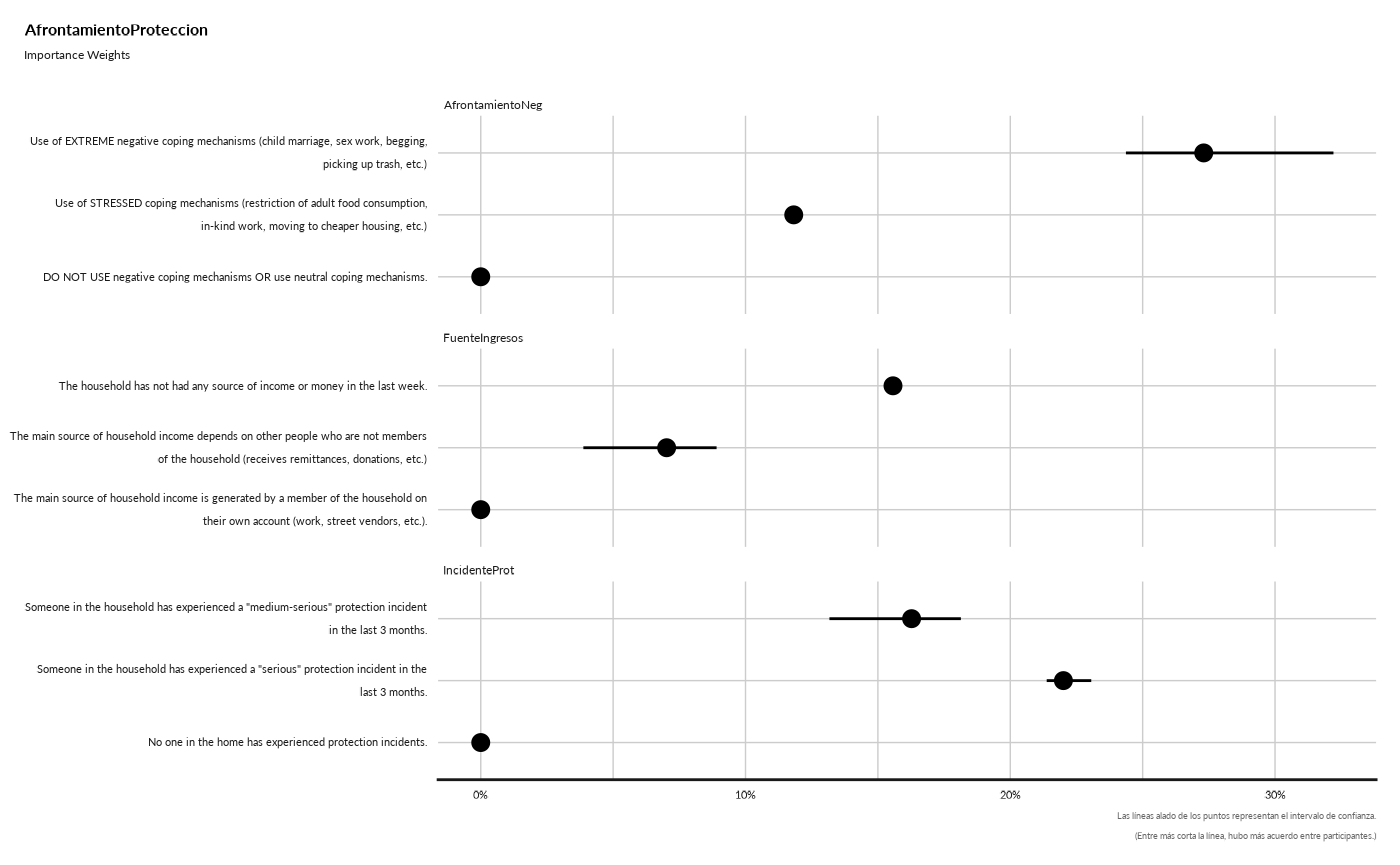
## Save a csv extract of the weights
# purrr::walk2(cjdata$dim, cjdata$amces, ~write_csv(.y, fs::path(.x, ext = "csv")))